The EEGLAB News #3
ICLabel Q&A
From: Scott Burwell <burwell@umn.edu>
Sent: Thursday, January 2, 2020 12:04 AM
To: eeglablist@sccn.ucsd.edu
Subject: Re: ICLabel: "source" explanation of "channel noise" independent
components in the absence of obviously bad data?
For background (to the uninitiated), the ICLabel plug-in returns a row of probabilities for each independent component that its source "class" is, respectively, brain, muscle, eye, heart, line noise, channel noise, or ‘other’ non-brain noise. Upon ICLabel returning source class probabilities for each component, there is a question as to how to go about filtering one's data to only "brain" components (e.g., for downstream scalp or source analyses). I have been testing out different cutoffs ... to keep only "brain" components, but have found this to feel a bit arbitrary (e.g., keep components with ‘brain’ probability > .90? > .75?). So, the approach I've taken lately is to designate a component's class to be the class for which its probability is the greatest (see below code snippet).
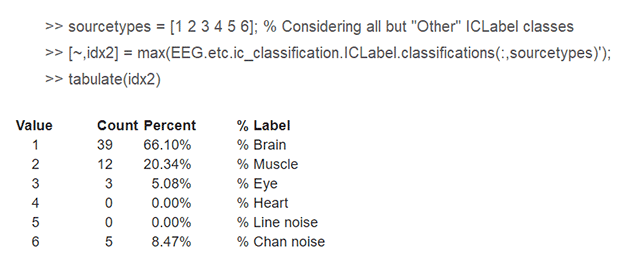
Thus, in this relatively clean looking resting-state dataset, ~50% of components are classified as "Brain," ~19% as "Muscle," and ~3% as "Eye," which upon my visual inspection appear to be accurate. Additionally, a substantial percentage of components are classified as "Channel noise" (~5%) and "Other non-brain" (~22%), which I am finding difficult to explain / justify exclusion of in a manuscript I am writing, especially when the channel data appear to be clean. The percent variance accounted for by the "Channel noise" and "Other non-brain" components is small (<2%), but does not seem to be substantially different from the percent variance accounted for by components classified as "Brain." Additionally, the time-series activations and frequency spectra for the "Channel noise" and "Other" components do not appear to be terribly noisy or different next to that of some "brain" components.
I have thought about the possibility of only considering a *subset* of ICLabel columns in deciding their class. E.g., in the above code, specifying
>> sourcetypes = [1 2 3 4 5],
thereby effectively forcing "Channel noise" and "Other non-brain" components to be assigned to one of the easier to interpret classes (i.e., brain, muscle, eye, heart, line noise) -- but I am not sure how this would be received by others.
Your thoughts would be appreciated.
Scott Burwell
========================================================================
On Thu, Jan 2, 2020 at 1:44 PM Luca B Pion-tonachini <lpiontonachini@ucsd.edu> wrote:
The principal factor for determining a “Channel Noise” IC is the scalp topography. If the topography is very focal, that is often an indication of a “Channel Noise” component. All that really means is that the channel described by the IC is in some way already independent of the other channels prior to ICA decomposition. If the components ICLabel marked as “Channel Noise” look very focal (you could double check this by looking at the corresponding columns of the EEG.icawinv matrix: one element of each of those columns should have much higher magnitude), then ICLabel is labeling the ICs correctly and the real question is, “Why did ICA decompose those components that way?” If the components don’t actually look like they describe channel noise, then it is likely that ICLabel is wrong (I personally hope not, but it does happen), in which case you could state that as the likely explanation for the classification.
Luca Pion-Tonachini
========================================================================
From: *Scott Burwell <burwell@umn.edu>
Sent: *Friday, January 3, 2020 6:40 AM
To: *Luca B Pion-tonachini <lpiontonachini@ucsd.edu>
Thank you, Luca, for the clarification. Indeed, each topography of the components classified as "Channel Noise" is very focal, so perhaps the question I should ask is more of the ICA decomposition instead of the classification.
I am still a bit curious regarding how to handle components classified as "Other." Is there any unifying way to describe (and justify removal of) those components in a manuscript? In a large sample of subjects (n = 1500), I have found that the "Other non-brain" class is assigned nearly as often as the "Brain" class, the mean percentages of each classification type being: brain (37%), muscle (13%), eye (5%), heart (0%), line (2%), channel (8%), and other (35%).
Upon inspection of the "Other" class of components, the topographies do not look terribly messy or uninterpretable; rather, the topographies of the "Other" components look less ideal than the "Brain" components, but I am not certain this justifies excluding these components. I would like to avoid throwing out more dimensions of the data than what is needed, and 35% of components (in addition to the ~30% thrown out across Muscle, Eye, Heart, Line, and Channel Noise) seems like a lot of data.
I wonder how it would be received by others to only consider a subset of the columns in the ICLabel classifications output to "force" components classified as "Other" into one of the other class types? I.e., instead of considering all ICLabel classes / columns, consider only a subset of ICLabel classes / columns (1:6), forcing components above with the classification of "Other" into one of the classes that is (perhaps) better defined / easier to explain?
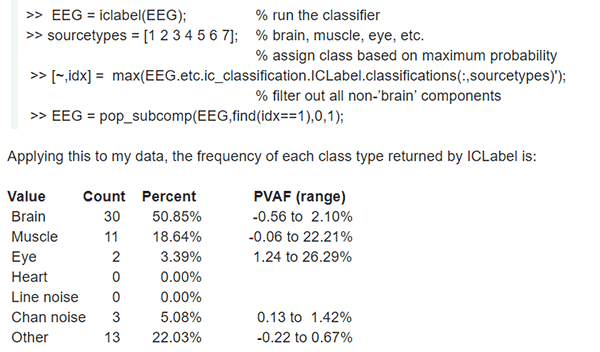
I am curious what you and others think about this approach? And what others have been doing for cutoffs / class selections with the classification probabilities?
p.s. Compliments on the crowd-sourced ICLabel classifier - it is a much needed tool and is very easy to run!
========================================================================
On Fri, Jan 3, 2020 at 11:07 AM Luca B Pion-Tonachini wrote:
“Other” is a catch-all for anything that does not fit ICLabel’s six other IC categories. Because that includes poorly-unmixed ICs, which may still contain significant brain activity, I highly suggest to *not* remove all “Other” ICs sight unseen. I thus stand in the more conservative camp of, “If you don’t know what it is, leave it alone.”
Regarding your idea of ignoring the “Other” category, I can’t say I’m a fan. I can understand doing that for specific components for which you disagree with the classification, but not all cases. If a component is actually purely noise, how does it make sense to call it “Brain” or “Eye” just because there was a 2% probability of that class? I think that your underlying purpose is to try to determine what is actually contained in the ICs classified as “Other” (correct me if I’m wrong) but I don’t think your can correctly do that with ICLabel without retraining the classifier on a dataset with a more detailed subcategorization of IC types.
Luca
========================================================================
Scott Burwell replied:
Thanks for your thoughts on how the "Other" category of component classifications should be interpreted / treated. I agree, if one doesn't know what a given component signifies, it is probably best to keep it until you have more corroborative evidence to exclude it.
I suppose if one were to determine whether to keep or remove components that were categorized as "Other," one could revert to more "classical" methods of filtering out components? E.g., keeping components with an equivalent current dipole / scalp map residual variance of less than some value (e.g., RV<15%), keeping components with a dipole location that is within a brain mask, etc.
Scott
========================================================================
Comment by Scott Makeig, Jan 6 7:47 PM:
To identify “Brain” ICs, applying the RV<15% threshold and “inside the brain” criterion, while heuristic, seem to work well (at least, across more or less equally clean datasets…). I am suggesting here to apply these criteria after selecting a pool of putative “Brain” ICs using ICLabel -- with or without forcing the “Other”-labeled ICs into another category, as you suggest.
Another dimension to think of is the relative strengths of the various ICs - particularly those classified by ICLabel as being in the “Other” category. How many of those ICs account for much data variance? This can be tested using eeg_pvaf() i, or guesstimated by considering the IC index (since ICs are typically returned in order of the data contributions). Thus, ICs with high indices typically make only quite small contributions to the data.
Incidentally, this observation applies still more to averaged ERP measures -- we typically find only 6-7 discernable IC contributors to (well-recorded, sufficiently averaged) ERPs (after removing large / distinct “Eye” ICs). And - these ERP-contributing ICs are typically those labeled as “Brain” by ICLabel and passing the (RV, in-brain location) quality tests above.
Much statistical analysis begins by attempting to separate the data into “signal” plus “noise” subspaces. ICA does not do this. However, the smallest (least contributing) ICs also tend to be those with ‘noisiest’ scalp maps, and are seen to be the least stable under repeated decompositions of the data (e.g., as in RELICA). Hence, they can be thought of as constituting a relatively small noise subspace of the data identified by ICA.
But note: the ‘unreliability’ of some ICs reported by RELICA just might reflect some fundamental nonstationarity in the data (for example: Did the subject nod off during the experiment, and were some of the ICs active only in those periods?). We are now (re)examining the power of AMICA to separate data into stationary periods, and hope to publish tools for evaluating what it reveals in the coming year.
Scott
p.s. In Scott Burwell’s first table at the top of this email thread, note that some returned PVAF (percent variance accounted for) values for some ICs are <0% How can this be? This is because PVAF answers the question, “How much smaller are the data once you have subtracted out the putative portion or model of the data?” If what you subtract is wildly unlike the data themselves, then the result may actually be bigger than the original data (and hence, PVAF < 0%)!