The EEGLAB News #3
NSG-enabled RELICA plug-in
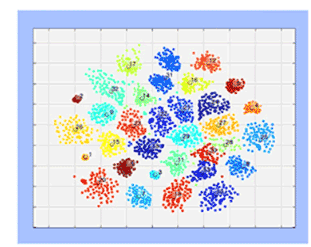 RELICA (Artoni et al., 2014) now runs faster - or much faster still! RELICA, for ‘RELiability of ICA’, uses a stochastic approach to characterize the reliability of component processes identified by ICA decomposition of a multichannel dataset, by measuring the stability of the separated independent component processes under bootstrap resampling. To do this, RELICA performs repeated ICA decompositions of bootstrapped versions of the input data, then measures the stability of the returned ICs. This process is highly computationally expensive, particularly for large datasets with many channels. (The figure shows RELICA IC replication clusters across multiple ICA decompositions of 100 bootstrap versions of an EEGLAB dataset, performed on NSG. Running time was 1/10 of the running time on a contemporary laptop.
RELICA (Artoni et al., 2014) now runs faster - or much faster still! RELICA, for ‘RELiability of ICA’, uses a stochastic approach to characterize the reliability of component processes identified by ICA decomposition of a multichannel dataset, by measuring the stability of the separated independent component processes under bootstrap resampling. To do this, RELICA performs repeated ICA decompositions of bootstrapped versions of the input data, then measures the stability of the returned ICs. This process is highly computationally expensive, particularly for large datasets with many channels. (The figure shows RELICA IC replication clusters across multiple ICA decompositions of 100 bootstrap versions of an EEGLAB dataset, performed on NSG. Running time was 1/10 of the running time on a contemporary laptop.
In the previous EEGLAB News issue, we announced the recent release of the EEGLAB plug-in nsgportal (github.com/sccn/nsgportal) that allows users to submit complex tasks to the Neuroscience Gateway (NSG) directly from within EEGLAB running on MATLAB on any lab computer. The plug-in nsgportal passes these jobs to NSG to run on the U.S. XSEDE network of high-performance computers (aka ‘supercomputers’). The iterative ICA process in the RELICA plug-in contributed to EEGLAB by collaborator Fiorenzo Artoni has, first, been parallelized using parfor loops from the MATLAB Parallel Computing Toolbox, which should speed processing to some extent on any current computer using multicore processors. In addition, we have now modified the plug-in by adding nsgportal code to enable it to run ten or more times faster still using NSG. Using the new version, the RELICA user can choose whether to run the computation locally or via NSG. The new RELICA is available from the EEGLAB Extension Manager, and from its GitHub repository (https://github.com/sccn/relica).
- Artoni, F., Menicucci, D., Delorme, A., Makeig, S., Micera, S. “RELICA: A method for estimating the reliability of independent components.” NeuroImage 103:391-400, 2014.