The EEGLAB News #4
 Benedikt Ehinger
Benedikt Ehinger
Donders Institute for Brain, Cognition and Behaviour in Nijmegen, The Netherlands
Psychology, Postdoctoral fellow, https://benediktehinger.de
Olaf Dimigen
Humboldt-Universität zu Berlin, Germany
Visiting professor for Biological Psychology, http://olaf.dimigen.de
Sometimes inspiration hits at just the right time. That was the case for Drs. Benedikt Ehinger (Postdoctoral researcher at Donders Institute for Brain, Cognition and Behaviour in Nijmegen, The Netherlands) and Olaf Dimigen (Visiting professor at Humboldt-University in Berlin, Germany). Over coffee at a scientific conference in Vienna, in 2015, they found out that they both struggled with the analysis of temporally overlapping brain potentials from eye movements. After some discussion, they decided to write an open source toolbox that allows researchers to separate such overlapping EEG signals using deconvolution, a technique used to enhance signals from recorded data.
The Unfold Toolbox
Drs. Ehinger and Dimigen both work with electroencephalography (EEG) – monitoring the electrical activity of the brain supporting thought and action via the fraction of its potentials that reach the scalp. Dr. Ehinger explains: "If we want to record the EEG in naturalistic situations – for example while reading a book, looking at a piece of art, or walking around in a (possibly virtual) city – one problem is immediately apparent: Many things happen at nearly the same time. The recorded EEG typically sums brain activity associated with complex interleaved sensory inputs and motor action events. However, even in traditional, well-controlled laboratory experiments, EEG measures typically reflect the overlapping activities associated with multiple events during the trial, such as stimulus onsets, involuntary microsaccades, and button presses. Such overlap can both produce confounding effects or obscure real effects. If we want to separate these potentials and move towards more ecologically valid experimental designs, we need a different analysis approach: deconvolution based on multiple linear regression. Whereas independent component analysis (ICA) separates the spatially overlapping activity of different sources, deconvolution separates temporally overlapping activity."
 Upon first review of relevant literature, Drs. Ehinger and Dimigen realized they were not the first to come up with this idea. Just in the same year, a twin-paper by Smith & Kutas (2015) came out in Psychophysiology. "And the fMRI community embraced this approach quite extensively already in the 1990s," Dr. Dimigen explains. "It was also used in EEG. For example, in Dr. Makeig’s lab, they were making the first prototypes of this approach available in 2013." And it was perhaps first introduced in one form (ADJAR) by Marty Woldorff at UCSD in the early 1980s.
Upon first review of relevant literature, Drs. Ehinger and Dimigen realized they were not the first to come up with this idea. Just in the same year, a twin-paper by Smith & Kutas (2015) came out in Psychophysiology. "And the fMRI community embraced this approach quite extensively already in the 1990s," Dr. Dimigen explains. "It was also used in EEG. For example, in Dr. Makeig’s lab, they were making the first prototypes of this approach available in 2013." And it was perhaps first introduced in one form (ADJAR) by Marty Woldorff at UCSD in the early 1980s.
But after becoming more familiar with these efforts, Drs. Ehinger and Dimigen noticed a lack of documentation and usability: "We saw the need for a tool that was more intuitive to use and that allows us to simulate data and thoroughly test whether and where the approach is useful." So they made it their goal to create that tool. The final product: “Unfold: an integrated toolbox for overlap correction, non-linear modeling, and regression-based EEG analysis.”
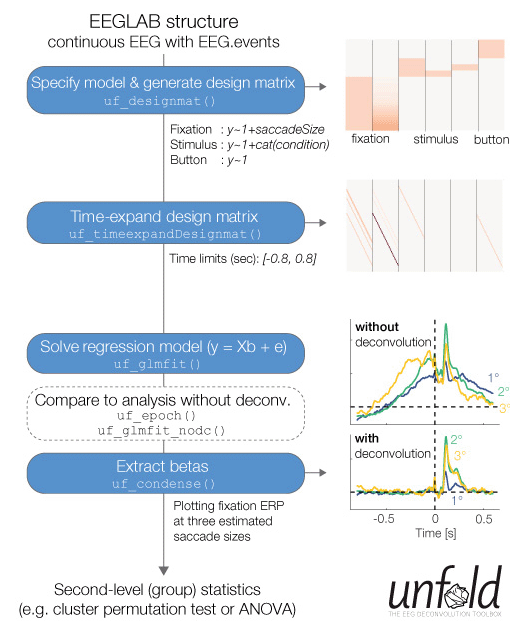
There are three reasons why Unfold is easy to adopt into an EEGLAB workflow: (1) Unfold makes use of the popular Wilkinson Formulas (used in R, Python, and Matlab statistics) to specify linear models, (2) the Unfold documentation has many tutorials of increasing complexity to help you get started, and (3) Unfold uses the EEGLAB data structure with which many users are already familiar. (Figure to right: Overview of typical analysis steps in Unfold applications.)
Current Use
"Now, five years later, we can conclude that the approach works really well”, Dr. Dimigen remarks, “for example when we want to model eye movement-related brain responses in active viewing situations like scene perception or reading." Drs. Ehinger and Dimigen both apply the Unfold toolbox extensively in their work. For example, in a recently submitted paper, the two analyzed combined eye-tracking/EEG experiments with (non)linear deconvolution models.
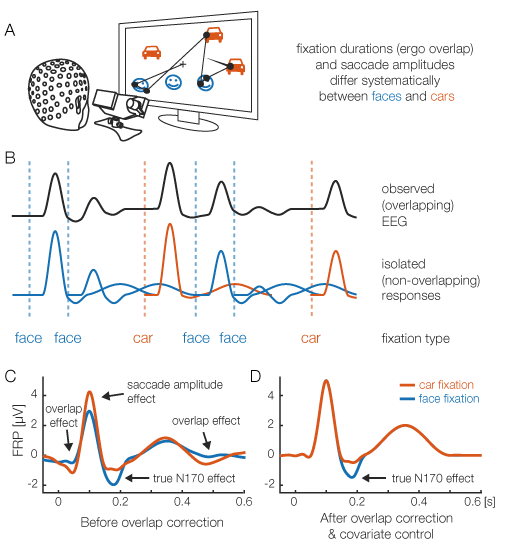 (Figure to left: (A) The concept of deconvolution, illustrated for simulated data of an eye-tracking/EEG experiment in which participants look at faces and cars. (B) A short interval of the recorded EEG. Every eye fixation elicits its own brain response (lower row). However, because brain responses last much longer than a typical fixation duration, we can only observe the sum of overlapping responses (upper row). (C) Average fixation-related brain potential (FRP) of each condition. In addition to the genuine N170 effect of looking at a face, this simulation reveals several spurious effects, caused by the systematic differences in fixation duration and saccade size between the two conditions. Here we assume shorter fixation durations for faces, and thus the overlap with the next fixation is stronger. We also simulate a difference in saccade size between conditions. (D) Deconvolution corrects for both spurious effects. Now we can recover the true N170 effect of looking at a face, free of confounds.) (adapted from Dimigen & Ehinger, "Analyzing combined eye-tracking/EEG experiments with (non) linear deconvolution models," bioRxiv, 2019).
(Figure to left: (A) The concept of deconvolution, illustrated for simulated data of an eye-tracking/EEG experiment in which participants look at faces and cars. (B) A short interval of the recorded EEG. Every eye fixation elicits its own brain response (lower row). However, because brain responses last much longer than a typical fixation duration, we can only observe the sum of overlapping responses (upper row). (C) Average fixation-related brain potential (FRP) of each condition. In addition to the genuine N170 effect of looking at a face, this simulation reveals several spurious effects, caused by the systematic differences in fixation duration and saccade size between the two conditions. Here we assume shorter fixation durations for faces, and thus the overlap with the next fixation is stronger. We also simulate a difference in saccade size between conditions. (D) Deconvolution corrects for both spurious effects. Now we can recover the true N170 effect of looking at a face, free of confounds.) (adapted from Dimigen & Ehinger, "Analyzing combined eye-tracking/EEG experiments with (non) linear deconvolution models," bioRxiv, 2019).
The Unfold toolbox is being used by many researchers and being applied in diverse contexts. Dr. Ehinger rattles off an impressive list of current applications, including: "Reading and spoken language comprehension, anti- & prosaccade tasks in aging, navigation in virtual environments, face perception inside and outside the laboratory, mobile EEG, evidence integration in a P300 task, heart beat potentials and pain, visual search with pointing movements of the arm, or infant research."
The Future
Both researchers agree that the toolbox appeared at a good time. "Many EEG and MEG groups are currently starting to use deconvolution," Dr. Dimigen shares, "so we are lucky to have early adopters that provide us with feedback on usability and features."
"We have also recently added a number of new features," Dr. Ehinger adds. "These include functions for cross-validation, for plotting ERP-images of partial effects, and for computing the temporal response functions (TRFs) of time-continuous predictors (like the sound envelope of a music track). More functions are in the pipeline."
And both researchers are enthusiastic about a new, more exploratory direction -- connected to a new, sister toolbox, 'Unmixed'. This extension to Unfold uses mixed linear models to calculate single-subject estimates of deconvolved brain responses. "It would also be useful to include a simple graphical user interface, so models can also be run directly from the EEGLAB menus, instead of only from the MATLAB command line."
Drs. Ehinger and Dimigen are full of ideas for future applications in their own projects. While they don't know exactly where and how far their cutting-edge method will lead them, one thing is clear: They are ready to bring their best tools on their journey. Even, or especially, if it means making their own – and then sharing them with other researchers.
R. Weistrop, April 2020