The EEGLAB News #4
 Kay Robbins, Ph.D.
Kay Robbins, Ph.D.
Professor Emeritus, Department of Computer Science,
University of Texas at San Antonio
http://www.cs.utsa.edu/~krobbins/
Visualization and Modeling Laboratory: http://visual.cs.utsa.edu/
"I became interested in electroencephalography (EEG) when I met Scott Makeig and had the opportunity to join a large collaborative grant about Neuroergonomics that he was leading, more than a decade ago. I found the idea of Neuroergonomics – of measuring human performance and the brain in the real-world – absolutely compelling, and have been working in that area ever since.”
Kay Robbins, Ph.D., Professor Emeritus, Department of Computer Science, University of Texas at San Antonio, describes her first encounter with Neuroergonomics, and how it immediately captured her attention and altered the course of her research. The field is exciting because it brings neuroscience into everyday life. She explains, “This research will provide a new and data-intensive glimpse of life in an event-rich world. We hope that a better understanding of the interaction between brain and body during real-world behavior will lead to technology for monitoring attention and fatigue as well as reducing cognitive load for people operating in normal and high-stress conditions.”
Dr. Robbins heads The Visualization and Modeling Laboratory (VML) at the University of Texas at San Antonio. Her goal: “To understand the world through data.” Her lab integrates visualization, machine learning, statistical analysis, various modeling techniques with usable interfaces, and management of large scale, multimedia data sets to look at problems in new ways. The research team currently focuses on tools to measure the brain and body at work, with data from EEG, eye-tracking, motion capture and the environment.
EEGLAB
Dr. Robbins is a strong enthusiast of EEGLAB. She uses the toolbox for many aspects of her work, and is an avid tool developer. “I am a big fan of open-source science,” she shares emphatically. “We write a lot of scripts and tools in my lab. Recently, we called EEGLAB from python as part of the front-end processing for using deep learning networks on EEG.”
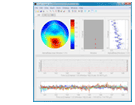
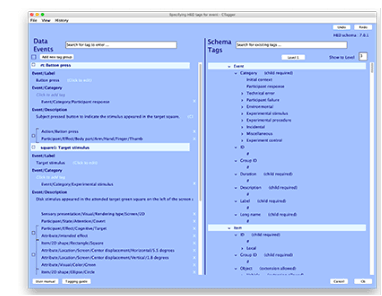 She and her students have developed a long list of open source EEGLAB-plugins, including “the PREP Pipeline, a standardized preprocessing pipeline for EEG data, the HED (hierarchical event descriptors) tagging system for identifying experimental events, along with the CTAGGER tool set for managing tags, and BLINKER a tool for automatically detecting and annotating blinks in EEG data.” (see the full list of plug-ins developed in her lab, below. Image to right: HED interface)
She and her students have developed a long list of open source EEGLAB-plugins, including “the PREP Pipeline, a standardized preprocessing pipeline for EEG data, the HED (hierarchical event descriptors) tagging system for identifying experimental events, along with the CTAGGER tool set for managing tags, and BLINKER a tool for automatically detecting and annotating blinks in EEG data.” (see the full list of plug-ins developed in her lab, below. Image to right: HED interface)
And Dr. Robbins is eager to share these tools with others – part of her strong belief in open-source science. “We focus on building tools that are usable not only in our lab,” she adds, “but for other researchers, as well.”
Early Years
How did someone growing up in a small town in Wisconsin become interested in large scale, multimedia data sets? “Somehow,” she says laughing, “I managed to make it to MIT for both undergraduate and graduate work.” There, she earned degrees in mathematics (dynamical systems, modeling, fluid dynamics). From her earliest research, Dr. Robbins was interested in trying to extract meaning and structure directly from experimental data. She realizes that working with large scale, multimedia data sets gives researchers the ability to look at problems in new ways.
Swartz Center for Computational Neuroscience
Dr. Robbins thrives in a collaborative environment. “Over the years, I have had long-term collaborations with several researchers in other fields,” she explains. “I have done work in many areas including dynamical systems, fluid dynamics, combustion, imaging and bio-informatics – always partnered with wonderful experimental colleagues.” In fact, that is how she met Scott Makeig at the Swartz Center for Computational Neuroscience (SCCN) – as a collaborator on the large grant, “Neuroergonomics: Studying the Brain and Body at Work." From that fateful partnership beginning, a decade ago, she developed a fascination with measuring human performance in the real-world, which hasn’t stopped!
Dr. Robbins eagerly describes other connections she has with SCCN researchers: “I have collaborated with several people who were at SCCN at the time (Nima Bigdely-Shamlo, Tim Mullen, and Christian Kothe) for some of this work. And in terms of plug-ins, I am collaborating closely with Scott Makeig, Dung Truong, and Arno Delorme on the further development of HED so that it can be incorporated into EEGLAB and included as part of their new computational portal NEMAR. HED, which has become the de facto brain imaging data event annotation standard for the community.”
 When Dr. Robbins mentions ‘NEMAR”, she is referring to an ongoing five-year, nearly $5 million grant from the U.S. National Institutes of Health (NIH) to create a portal (‘NEMAR’) for human electrophysiological data. This portal will be offered through OpenNeuro (a free and open platform for sharing MRI, MEG, EEG, iEEG, and EcoG data) developed by Russ Poldrack and colleagues at Stanford University. SCCN’s Scott Makeig and Arnaud Delorme, with co-Pis Amitava Majumdar from UC San Diego’s San Diego Supercomputer Center (SDSC), and Russ Poldrack of Stanford University were awarded the NEMAR grant in August 2019. This step could have an increasing influence on the future of brain research.
When Dr. Robbins mentions ‘NEMAR”, she is referring to an ongoing five-year, nearly $5 million grant from the U.S. National Institutes of Health (NIH) to create a portal (‘NEMAR’) for human electrophysiological data. This portal will be offered through OpenNeuro (a free and open platform for sharing MRI, MEG, EEG, iEEG, and EcoG data) developed by Russ Poldrack and colleagues at Stanford University. SCCN’s Scott Makeig and Arnaud Delorme, with co-Pis Amitava Majumdar from UC San Diego’s San Diego Supercomputer Center (SDSC), and Russ Poldrack of Stanford University were awarded the NEMAR grant in August 2019. This step could have an increasing influence on the future of brain research.
Looking to the Future
The field of data science today is a far cry from where it was when Dr. Robbins began: “Before digital computers, we used analog computers to perform simulation experiments. Over the years much of the struggle has been to get more data, more memory, and faster computation.” At that time, public access to data was very limited. But now, she explains, “the issues have changed, and I must say, it is a relief.”
With these changes, Dr. Robbins has found a renewed sense of focus. “I am a firm believer in reproducible science. This means not only publishing data and code, but also using automated techniques, conducting same subject replications, and developing easy-to-use analysis tools for assessing data variability.” She continues, “I think that HED is an important step in making this possible, as it allows for annotation of events using a common vocabulary so that automated computation is possible. I also think that making sure that well-curated data is publicly available is very important. Data curation is not very glamorous but I think it is vital to progress. The public availability of EEG data has been extremely limited, but now BIDS has standardized formats for EEG and openNeuro is accepting EEG data, so some progress has been made.”
With the latest technological advances in the field and the potential for improved access to EEG data, Dr. Robbins envisions a bright future. “I hope that NEMAR will provide a development platform to make it easier to do good science -- I'm, working to see that this happens.”
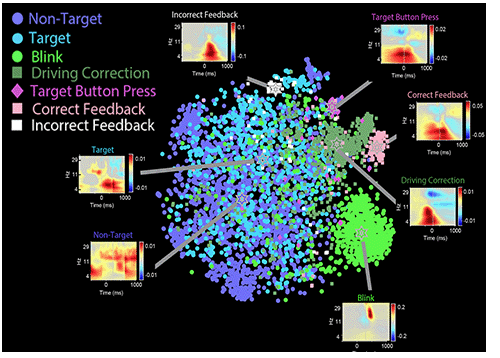 Figure from a recent UTSA Visualization and Modeling Lab (VML) paper. 2D t-SNE visualization of rERSPs for 26 channels in common across 17 studies with different experimental designs and stimuli. Each point represents a vector formed by the rERSPs concatenated over all channels and frequencies for the interval [−1, 1] seconds corresponding to a study-specific event code for one recording. Points are color-coded by the most prominent HED tag associated with the event code. The median coordinates of points associated with a particular HED tag are marked with a star. Inset plots of the average of the rERSPs of all points associated each HED tag are shown for channel FCz. The scale for these plots is fraction of the median signal, with gray denoting non-significance. The analysis finds commonalities in event HED tag descriptors across studies. Note the beta bursts (seemingly unique in these data) following eye blinks across experiment designs (lower right),
and the overlapping (in this visualization) ERSP features associated with target versus nontarget stimulus presentations (left two ERSP features).
Note also the highly distinguishable occurrences of EEG patterns associated with Correct and Incorrect feedback presentations (top two ERSP features). Bigdely-Shamlo, N., Touryan, J., Ojeda, A., Kothe, C., Mullen, T., and Robbins, K. (2019). Automated EEG mega-analysis II: Cognitive aspects of event related features, Neuroimage. 2019 Sep 4:116054. doi: 10.1016/j.neuroimage.2019.116054, PMID: 31491523.
Figure from a recent UTSA Visualization and Modeling Lab (VML) paper. 2D t-SNE visualization of rERSPs for 26 channels in common across 17 studies with different experimental designs and stimuli. Each point represents a vector formed by the rERSPs concatenated over all channels and frequencies for the interval [−1, 1] seconds corresponding to a study-specific event code for one recording. Points are color-coded by the most prominent HED tag associated with the event code. The median coordinates of points associated with a particular HED tag are marked with a star. Inset plots of the average of the rERSPs of all points associated each HED tag are shown for channel FCz. The scale for these plots is fraction of the median signal, with gray denoting non-significance. The analysis finds commonalities in event HED tag descriptors across studies. Note the beta bursts (seemingly unique in these data) following eye blinks across experiment designs (lower right),
and the overlapping (in this visualization) ERSP features associated with target versus nontarget stimulus presentations (left two ERSP features).
Note also the highly distinguishable occurrences of EEG patterns associated with Correct and Incorrect feedback presentations (top two ERSP features). Bigdely-Shamlo, N., Touryan, J., Ojeda, A., Kothe, C., Mullen, T., and Robbins, K. (2019). Automated EEG mega-analysis II: Cognitive aspects of event related features, Neuroimage. 2019 Sep 4:116054. doi: 10.1016/j.neuroimage.2019.116054, PMID: 31491523.
Open Source EEGLAB-plugins developed by Dr. Robbins and her colleagues/students at VML (for longer descriptions, click here):
 The PREP Pipeline, a standardized preprocessing pipeline for EEG data, that focuses on the identification of bad channels and the calculation of a robust average reference.
The PREP Pipeline, a standardized preprocessing pipeline for EEG data, that focuses on the identification of bad channels and the calculation of a robust average reference.
- The HED (hierarchical event descriptors) tagging system for identifying experimental events;

- HEDTools - a MATLAB/Java Toolbox and an EEGLAB plugin designed to help users annotate and validate events or other data elements using a predefined, but extensible, hierarchically structured annotation language.
- The HED Validator - an online-tool designed to make validation of HED tags easy.
- The CTAGGER tool set for adding and managing HED event tags;
 BLINKER - a tool for automatically detecting and annotating blinks in EEG data,and calculating variousproperties of these blinks;
BLINKER - a tool for automatically detecting and annotating blinks in EEG data,and calculating variousproperties of these blinks;
- DETECT - a MATLAB Toolbox for detecting and identifying events in long multi-channel time series;
- EEG-Annotate - A tool to identify a given response in new continuous EEG signals;
 EEG-HDF5-Tools - Conversion utilities for EEG using HDF5. Tools are provided for Java, C, R, Python, and MATLAB;
EEG-HDF5-Tools - Conversion utilities for EEG using HDF5. Tools are provided for Java, C, R, Python, and MATLAB;
- EEGVIS - a MATLAB toolbox that allows users to quickly explore multi-channel EEG and other large array-based data sets using multi-scale drill-down techniques;
- MOBBED - a lightweight, easy-to-use, extensible toolkit that allows users to incorporate
 a computational database into their normal MATLAB workflow.
a computational database into their normal MATLAB workflow.
R. Weistrop, March 2020