The EEGLAB News #7
From the eeglablist
Q: Justin Fine, Post-doctoral researcher at Indiana University: I have a question that the literature does not really seem to answer regarding source analysis: Why should someone prefer to use a given source method to find EEG sources in areas that are known or assumed a priori? Specifically, I am asking about the benefits and obvious trade-offs of (1) an ICA + equivalent current dipole (ECD) modeling, else (2) distributed (sparse or group) hierarchical source localization methods (e.g., MSP in SPM) or, (3) a Bayesian ECD approach that does not rely on fitting separate IC components but rather on specifying source locations a priori?
Quick background: I have T1 MR head images and recorded electrode positions (64-channel Acticap) for all participants. The main goal here is to extract time-frequency measures and evoked potentials (ERPs) from sources in three cortial areas: rIFG, pre-SMA/ACC/MCC, and left M1. The study is a standard stop signal task, for which the literature tends to prefer the method (1) using ICA + ECD fitting. But I gather that might have something to do with researchers typically (1) not having electrode locations and (2) not having T1 head images? Any thoughts and feedback would be greatly appreciated. Thanks!
A: Scott Makeig: Justin - You are right that the choice of inverse source localization approach typically depends on the information available as well as the goals of the analysis. If you have T1 images for your study participants, I assume you also measured the 3D locations of the electrodes? If so, I suggest you try using the NFT and NIST toolboxes by Zeynep Akalin Acar, as these build electrical head models, optionally optimize these models using the SCALE algorithm for estimating the individual skull conductance (the largest unknown affecting EEG source localization), and then perform either (optimized) equivalent dipole localization OR high-resolution distributed source localization using the SCS 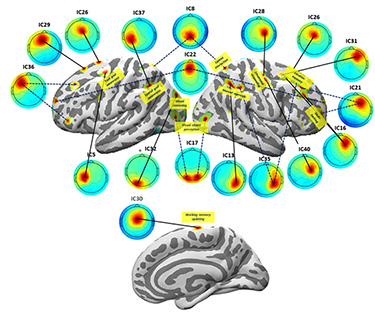 (Sparse-Compact-Smooth) localization algorithm of Cheng Cao.
(Sparse-Compact-Smooth) localization algorithm of Cheng Cao.
(Figure: SCALE/SCS-estimated source distributions and matching whole-data IC scalp maps, connected by line segments, for 17 of 45 brain IC effective sources drawn from a single-model AMICA decomposition of 128-channel data recorded during a complex (STRUM) multiscreen video game playing task session, superimposed on the participant’s semi-inflated cortical surface mesh constructed using the NFT toolbox from the participant MR head image. Dashed (not solid) connecting line segments show IC source estimates again using a SCALE-optimized head model by SCS but consisting of two (not one), presumably anatomically well-connected source areas. Labels indicate area functional associations in the functional imaging literature. From: Z. Akalin Acar & S. Makeig, IEEE BIBE, 2020. Click on the figure for an enlarged view.)
To extract all the spatial source information possible from the individual records, one must use optimized, computationally intensive methods. Because of individual differences in cortical folding, group source analysis limits spatial resolution somewhat; even high-resolution localization performed on sources in individual brains then leaves the problem of finding equivalent sources across individuals (i.e., equivalent source clusters). The best approach I know of is that of Arthur Tsai (Tsai et al., 2014) based on advanced cortical co-registration in Freesurfer. So far as I know, no open source software to accomplish this has yet been released.
But in considering different methods, one must first think clearly about how one defines an EEG 'source' -- that is, ‘a single’ EEG source.... What identifies a source as one functional source? Certainly, any source resolvable from the scalp data must represent coherent field potential across a large (>cm2) patch of cortex. A raw scalp channel represents a net spatially-weighted difference in local field potentials across more than one broad cortical territory, as Zeynep's CRF ('cortical receptive field') function (to be released soon, I hope) clearly shows.
For example, does the scalp map at a peak in a trial-mean ERP (or other EEG measure) represent the projection of "a source" (that is, "one single source")? It does if that is how you define "source" (!) -- but that definition may have limited neuroscientific value, as this "source" may actually represent the sum of several "local cortical source" activities in several cortical areas, with different functional significances -- as ICA decomposition, followed by source localization often reveals.
The neuroscientific value of ICA decomposition, I believe, is that it finds projections of single (or dual, highly-connected) cortical 'patches' within/across which local field potential is relatively coherent across a sufficient portion of the decomposed data. These 'source patch' projections should dominate scalp recordings, as local field activities in other cortical areas that are spatially incoherent will be largely cancelled out in the scalp channel signals by phase cancellation. If they do dominate the net scalp signals, then other inverse source estimation approaches should also locate them, at least approximately...
I hope these thoughts might be helpful. - Scott
p.s. Justin wrote further to ask whether there are yet any quantitative 'ground truth' studies (based on simulated phantom heads, concurrent invasive recording or MEG, etc.) that compare different methods of source analysis including ICA-based. Unfortunately, I know of none to date, but would be happy to collaborate on such studies.