FMRLAB Tutorial
v 2.0
v 2.0
©Jeng-Ren Duann & Scott Makeig, 2002
Swartz Center for Computational Neuroscience
Institute for Neural Computation
University of California San Diego
Swartz Center for Computational Neuroscience
Institute for Neural Computation
University of California San Diego
4. Image Preprocessing
To create an FMRLAB dataset: (1) Select a functional
image file. (2) Enter the image acquisition parameters for the functional
scans. (3) Enter the image information for the structural scans.
Select Dataset > Create Dataset from the
FMRLAB menu. A "Select Image File"
window will pop up (as below) allowing
you to select the file containing the functional scans. For the example dataset,
select "2dseq_r1"
as the functional image file.
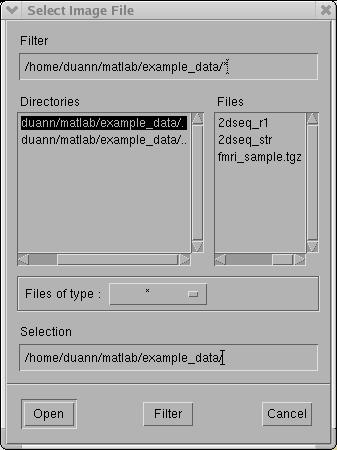
Move the mouse cursor over "2dseq_r1"
(the example functional dataset) and click the "Open"
button at the bottom
of the window. Next, a "Functional Image Information"
window will pop up (as below) allowing you to enter the image acquisition
parameters for the functional scans.
Fill all the fields with the values given in
Section 2.5 aobve.

After filling in the correct values in all the fields, press the
"OK" button
at the bottom of the window. Next, the
"Structural Image Information"
window will pop up (as below) to allow you to enter the necessary
structural image parameters.
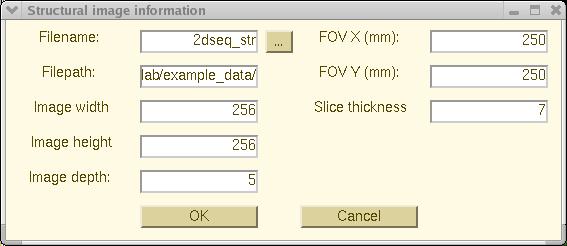
Press the (top center) continuation button [...]
and a "File Selection"
dialog will pop up to allow you to select the structural image file. In
this example, please select file "2dseq_str".
Again refer to the structural image acquisition parameters
(Section 2.5 above) to enter the correct
values in the other fields of this window.
4.2 Save the FMRLAB dataset
To save the dataset you have just created, select Dataset > Save Dataset from the FMRLAB menu. A file
selection window will pop up allowing you to input the output filename. Be
sure to append the extension .fmr
to the output dataset file. Then,
click the Save button to save the dataset
to disk.
4.3 Remove dummy scans
You specified the number of initial "dummy" scans during dataset creation
(see Section 4.1 above). To remove
those scans from the fMRI time series, select Process >
Remove Dummies from the menu.
4.4 Slice timing adjustment
To adjust the slice timing (using interpolation to make the
acquisition times for the slices as synchronous as possible), select
Process > Slice Timing. A
"Slice Timing Window"
will pop up (as below) allowing you to specify the sequence in which the
functional slices were acquired. There are four possible selections:
Interleaved, Ascending, Descending and
User Defined. The default is
Interleaved.
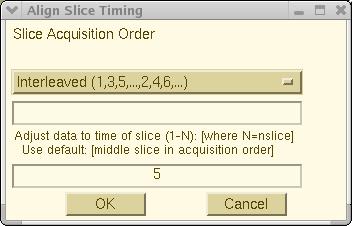
To use User Defined mode, enter the actual slice
acquisition sequence of the run, for example,
"2 4 6 8 10 1 3 5 7 9", into the text entry field
of the "Slice Timing Window".
This window also allows you to specify which
slice the slice timing adjustment should be aligned to. Simply give a slice
number in the editing field at bottom portion of this window. Else, the slice
number in the middle of the specified slice acquisition order will be
used as the default. Then, press OK
to start the slice timing adjustment process.
During this process, a progress-bar window will pop up to
indicate its progress. When the timing adjustment process is done, FMRLAB
will close both the Slice Timing Window
and the progress bar.
4.5 Remove off-brain voxels
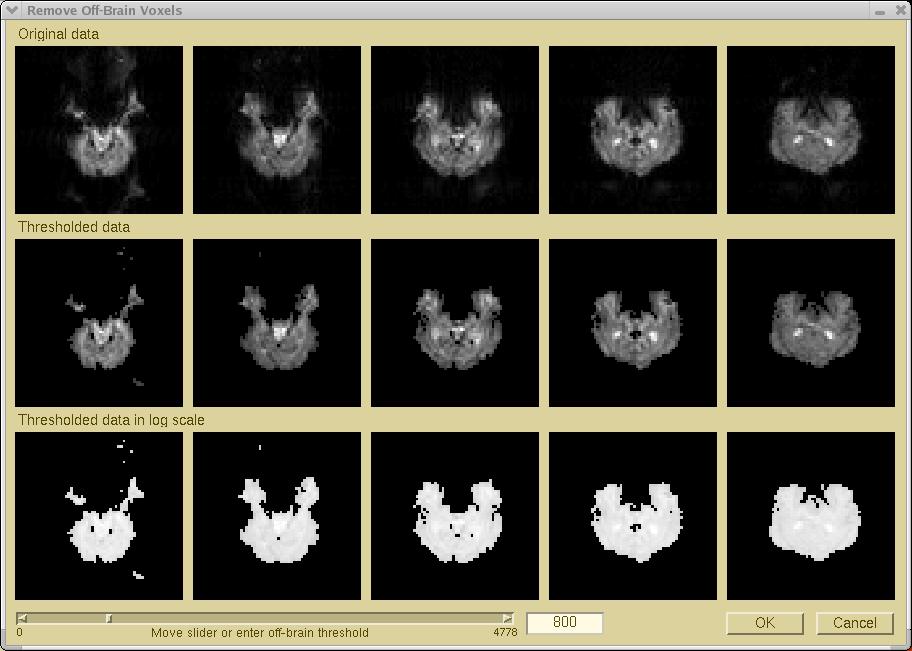
To remove off-brain voxels from the ICA training data, select Process > Extract Brain from the menu. The
"Remove Off-brain Voxels" window
will pop up. The first row will show
the original functional images (if the functional scans include more
than five slices, the middle five slices will be used for this display).
The middle row will show the functional images after thresholding,
and the bottom row, the log-transformed images after thresholding.
Because initially the threshold (displayed at the bottom) is
"0", at first there will not be any difference between
the top and middle rows.

To adjust the threshold, either mouse-click or drag the scoll
bar, else enter a numeric off-brain threshold value in the editing field
at the bottom of this window. When the threshold is changed, the display window
will be refreshed. You may update the threshold interactively until the
off-brain voxels in the functional images are removed as cleanly
as possible. At the same time, take care not to remove voxels
in the brain region. Examining the log-scale images may help you to select
the appropriate threshold.
Press "OK" to accept the entered threshold
and to allow FMRLAB to manipulate the image matrix
appropriately for ICA decomposition.
Back to the table of contents ...
Next chapter: Decomposing the data with ICA ...
To create an FMRLAB dataset: (1) Select a functional image file. (2) Enter the image acquisition parameters for the functional scans. (3) Enter the image information for the structural scans.
Select Dataset > Create Dataset from the
FMRLAB menu. A "Select Image File"
window will pop up (as below) allowing
you to select the file containing the functional scans. For the example dataset,
select "2dseq_r1"
as the functional image file.

Move the mouse cursor over "2dseq_r1"
(the example functional dataset) and click the "Open"
button at the bottom
of the window. Next, a "Functional Image Information"
window will pop up (as below) allowing you to enter the image acquisition
parameters for the functional scans.
Fill all the fields with the values given in
Section 2.5 aobve.

After filling in the correct values in all the fields, press the
"OK" button
at the bottom of the window. Next, the
"Structural Image Information"
window will pop up (as below) to allow you to enter the necessary
structural image parameters.

Press the (top center) continuation button [...]
and a "File Selection"
dialog will pop up to allow you to select the structural image file. In
this example, please select file "2dseq_str".
Again refer to the structural image acquisition parameters
(Section 2.5 above) to enter the correct
values in the other fields of this window.
4.2 Save the FMRLAB dataset
To save the dataset you have just created, select Dataset > Save Dataset from the FMRLAB menu. A file
selection window will pop up allowing you to input the output filename. Be
sure to append the extension .fmr
to the output dataset file. Then,
click the Save button to save the dataset
to disk.
4.3 Remove dummy scans
You specified the number of initial "dummy" scans during dataset creation
(see Section 4.1 above). To remove
those scans from the fMRI time series, select Process >
Remove Dummies from the menu.
4.4 Slice timing adjustment
To adjust the slice timing (using interpolation to make the
acquisition times for the slices as synchronous as possible), select
Process > Slice Timing. A
"Slice Timing Window"
will pop up (as below) allowing you to specify the sequence in which the
functional slices were acquired. There are four possible selections:
Interleaved, Ascending, Descending and
User Defined. The default is
Interleaved.

To use User Defined mode, enter the actual slice
acquisition sequence of the run, for example,
"2 4 6 8 10 1 3 5 7 9", into the text entry field
of the "Slice Timing Window".
This window also allows you to specify which
slice the slice timing adjustment should be aligned to. Simply give a slice
number in the editing field at bottom portion of this window. Else, the slice
number in the middle of the specified slice acquisition order will be
used as the default. Then, press OK
to start the slice timing adjustment process.
During this process, a progress-bar window will pop up to
indicate its progress. When the timing adjustment process is done, FMRLAB
will close both the Slice Timing Window
and the progress bar.
4.5 Remove off-brain voxels
To remove off-brain voxels from the ICA training data, select Process > Extract Brain from the menu. The
"Remove Off-brain Voxels" window
will pop up. The first row will show
the original functional images (if the functional scans include more
than five slices, the middle five slices will be used for this display).
The middle row will show the functional images after thresholding,
and the bottom row, the log-transformed images after thresholding.
Because initially the threshold (displayed at the bottom) is
"0", at first there will not be any difference between
the top and middle rows.

To adjust the threshold, either mouse-click or drag the scoll
bar, else enter a numeric off-brain threshold value in the editing field
at the bottom of this window. When the threshold is changed, the display window
will be refreshed. You may update the threshold interactively until the
off-brain voxels in the functional images are removed as cleanly
as possible. At the same time, take care not to remove voxels
in the brain region. Examining the log-scale images may help you to select
the appropriate threshold.
Press "OK" to accept the entered threshold
and to allow FMRLAB to manipulate the image matrix
appropriately for ICA decomposition.

Back to the table of contents ...
Next chapter: Decomposing the data with ICA ...
